|
NANOTWEEZ |
|
Project type: PN-III-P4-ID-PCE-2016-0026
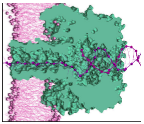
Description of the Project: Molecular recognition and binding mediated by weak non-covalent intermolecular interactions, are ubiquitous in many biological processes. Thus far, binding energy measurements rely on macroscopic approaches based on kinetic, saturation, or competition-modulation experiments, each of which are prone to specific advantages and shortcomings. As a new paradigm to studying molecular binding processes devoid of the drawbacks associated to existing techniques, we propose herein a single-molecule approach based on a protein nanopore sensor, to explore, probe and quantify the bond energy of non-covalent molecular complexes. In this regard, and since its inception over 20 years ago, nanopore-based analysis has evolved as a label-free, high-precision single-molecule approach for investigating biomolecular interactions and sensitive detection of molecules. Protein- and solid state-based nanopores have been established as powerful tools for the investigation of various chemistries or biomolecule detection and analysis, e.g., RNA and DNA, peptides and proteins. To alleviate critical shortcoming of current approaches and as a potential game-changer in the field of nanopore-based force spectroscopy, in this project we aim at exploring an approach to measure the binding energy of a duplex complex, at the single-molecule level, through capturing, retaining and then unzipping of a nucleic acid duplex inside an α-HL nanopore. As a state-of-the-art approach, we will use the 'nanopore tweezers' technique developed in our lab to capture and then reduce up to an almost stand-still of the escape rate of a tailor-functionalized biomolecule from a membrane immobilized α-HL.
Purpose of the project: We will develop the proof-of concept approach for the single-molecule nanomechanics, through applying controllable, oppositely oriented electric forces on a custom engineered, peptide-functionalized duplex biomolecule leading to its dissociation, and delivering in the end an estimate of the complex's bond energy. To demonstrate the utility of this approach, we will apply it to investigate the metal-mediated mismatched hybridization of natural DNA complexes, well-known in its implications but far less studied in molecular detail, with potential applications in nanotechnology as building block for self-assembling functional nanostructures.
Selected Objectives: ◊ Implementation, testing and validation of main protocols associated to the peptide-based, nanopore tweezer-force spectroscopy (NT-FS), for effective trapping and deforming of analytes in nanopores, with electric fields applied across the nanopore. ◊ Proof-of-concept of reproducible signature detection of polypeptide-functionalized ssDNA and dsDNA sequences with the α-HL nanopore. ◊ Deciphering molecular events reflecting electric field-mediated, polypeptide-functionalized dsDNA unzipping inside the α-HL nanopore. ◊ Thermodynamic description at uni-molecular level, of bond breakage and unzipping of dsDNA containing metal-mediated mispairs with the NT-FS technique. |

|
This project is funded under the National CDI Plan III UEFISCDI, as part of Program 4 - basic research and frontier, Exploratory Research (PCE) |


|
PN-III-P4-ID-PCE-2016-0026, nr. 33/12.07.2017 |
|
A nanopore tweezer-based approach for studying intermolecular interactions at uni-molecular level. Application to exploring metal-mediated, mismatched base pairs hybridization in nucleic acids (NANOTWEEZ) |